(Just from my point of view as a package maintainer.)
New users of R might not appreciate the full benefit of CRAN and new package maintainers may not appreciate the importance of keeping their packages updated and free of warnings and errors. This is something I only came to realize myself in the last few years so I thought I would write about it, by way of today’s example.
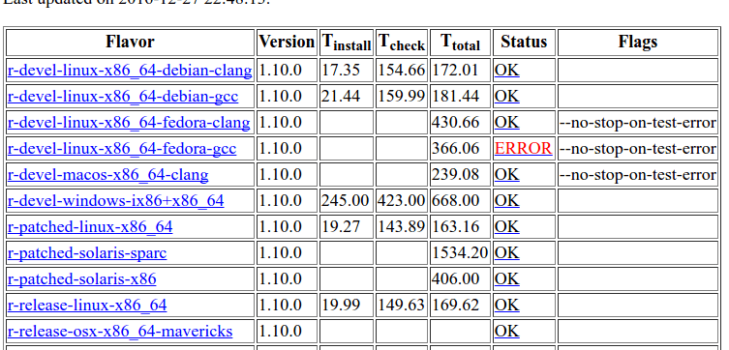
Since data.table was updated on CRAN on 3rd December, it has been passing all-OK. But today I noticed a new warning (converted to error by data.table) on one CRAN machine. This is displayed under the CRAN checks link.
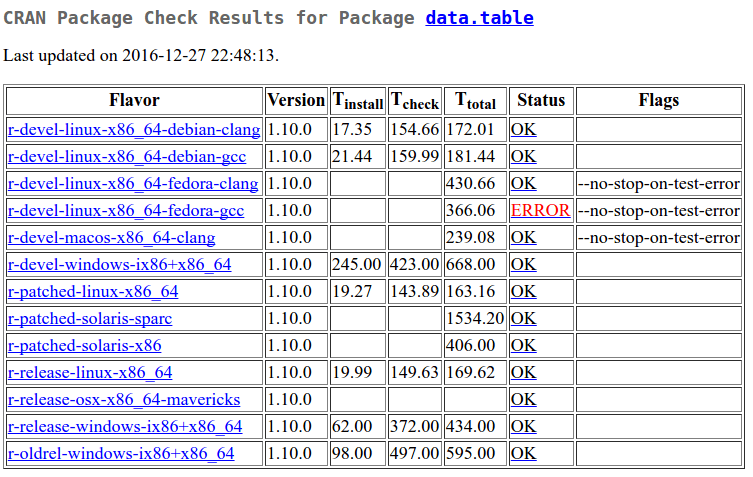
Sometimes errors happen for mundane reasons. For example, one of the Windows machines was in error recently because somehow the install became locked. That either fixed itself or someone spent time fixing it (if so, thank you). Today’s issue appears to be different.
I can either look lower down on that page or click the link to see the message.
Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
I’ve never seen this message before. But given it mentions something about something being deprecated and the flavor name is r-devel it looks likely that it has just been added to R itself in development. On a daily basis all CRAN packages are retested with the very latest commits to R that day. I did a quick scan of the latest commit messages to R but I couldn’t see anything relating to this apparently new warning. Some of the commit messages are long with detail right there in the message. Others are short and use reference numbers that require you to hop to that reference such as “port r71828 from trunk” which prevent fast scanning at times like this. There is more hunting I could do, but for now, let’s see if I get lucky.
The last line of data.table’s test suite output has been refined over the years for my future self, and on the same CRAN page without me needing to go anywhere else, it is showing me today:
5 errors out of 5940 (lastID=1748.4, endian==little, sizeof(long double)==16, sizeof(pointer)==8) in inst/tests/tests.Rraw on Tue Dec 27 18:09:48 2016. Search tests.Rraw for test numbers: 167, 167.1, 167.2, 168, 168.1.
The date and time is included to double check to myself that it really did run on CRAN’s machine recently and I’m not seeing an old stale result that would disappear when simply rerun with latest commits/fixes.
Next I do what I told myself and open tests.Rraw file in my editor and search for "test(167". Immediately, I see this test is within a block of code starting with :
if ("package:ggplot2" %in% search()) {
test(167, ...
}
In data.table we test compatibility of data.table with a bunch of popular packages. These packages are listed in the Suggests suggestion of DESCRIPTION .
Suggests: bit64, knitr, chron, ggplot2 (≥ 0.9.0), plyr, reshape, reshape2, testthat (≥ 0.4), hexbin, fastmatch, nlme, xts, gdata, GenomicRanges, caret, curl, zoo, plm, rmarkdown, parallel
We do not necessarily suggest these packages in the English sense of the verb; i.e., ‘to recommend’. Rather, perhaps a better name for that field would be Optional in the sense that you need those packages installed if you wish to run all tests, documentation and in data.table’s case, compatibility.
Anyway, now that I know that the failing tests are testing compatibility with ggplot2, I’ll click over to ggplot2 and look at its status. I’m hoping I’ll get lucky and ggplot2 is in error too.
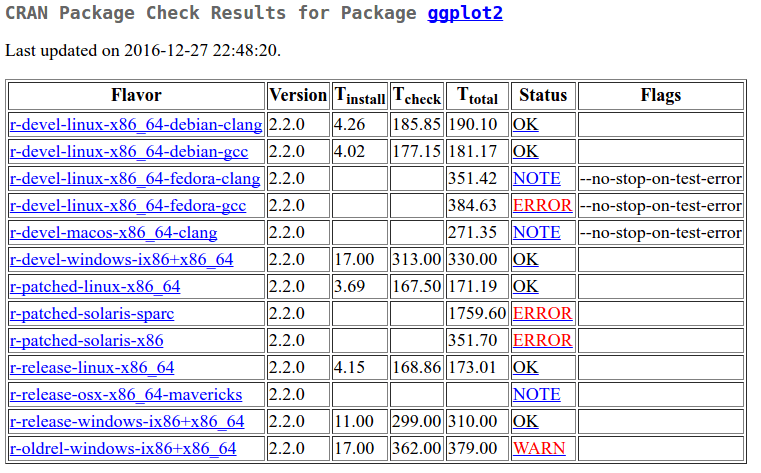
Indeed it is. And I can see the same message on ggplot2’s CRAN checks page .
Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
It’s my lucky day. ggplot2 is in error too with the same message. This time, thankfully, the new warning is therefore nothing to do with data.table per se. I got to this point in under 30 seconds! No typing was required to run anything at all. It was all done just by clicking within CRAN’s pages and searching a file. My task is done and I can move on. Thanks to CRAN and the people that run it.
What if data.table or ggplot2 were already in error or warning before R-core made their change? R-core members wouldn’t have seen any status change. If they see no status change for any of the 9,787 CRAN packages then they don’t know for sure it’s ok. All they know is their change didn’t affect any of the passing packages but they can’t be sure about the packages which are already in error or warning for an unrelated reason. I get more requests from R-core and CRAN maintainers to update data.table than from users of data.table. I’m sorry that I could not find time earlier in 2016 to update data.table than I did (data.table was showing an error for many months).
Regarding today’s warning, it has been caught before it gets to users. You will never be aware it ever happened. Either R-core will revert this change, or ggplot2 will be asked to send an update to CRAN before this change in R is released.
This is one reason why packages need to be on CRAN not just on GitHub. Not just so they are available to users most easily but so they are under the watchful eye of CRAN daily tests on all platforms.
Now that data.table is used by 320 CRAN and Bioconductor packages, I’m experiencing the same (minor in comparison) frustration that R-core maintainers must have been having for many years: package maintainers not keeping their packages clean of errors and warnings, myself included. No matter how insignificant those errors or warnings might appear. Sometimes, as in my case in 2016, I simply haven’t been able to assign time to start the process of releasing to CRAN. I have worked hard to reduce the time it takes to run the checks not covered by R CMD check and this is happening faster now. One aspect of that script is reverse dependency checks; checking packages which use data.table in some way.
The current status() of data.table reverse dependency checks is as follows, using data.table in development on my laptop. These 320 packages themselves often depend or suggest other packages so my local revdep library has 2,108 packages.
> status()
CRAN:
ERROR : 6 : AFM mlr mtconnectR partools quanteda stremr
WARNING : 2 : ie2miscdata PGRdup
NOTE : 69
OK : 155
TOTAL : 232 / 237
RUNNING : 0
NOT STARTED (first 5 of 5) : finch flippant gasfluxes propr rlas
BIOC:
ERROR : 1 : RTCGA
WARNING : 4 : genomation methylPipe RiboProfiling S4Vectors
NOTE : 68
OK : 9
TOTAL : 82 / 83
RUNNING : 0
NOT STARTED (first 5 of 1) : diffloop
Now that Jan Gorecki has joined H2O he has been able to spend some time to automate and improve this. Currently, the result he gets with a docker script is as follows.
> status()
CRAN:
ERROR : 18 : AFM blscrapeR brainGraph checkmate ie2misc lava mlr mtconnectR OptiQuantR panelaggregation partools pcrsim psidR quanteda simcausal stremr strvalidator xgboost
WARNING : 4 : data.table ie2miscdata msmtools PGRdup
NOTE : 72
OK : 141
TOTAL : 235 / 235
RUNNING : 0
BIOC:
ERROR : 20 : CAGEr dada2 facopy flowWorkspace GenomicTuples ggcyto GOTHiC IONiseR LowMACA methylPipe minfi openCyto pepStat PGA phyloseq pwOmics QUALIFIER RTCGA SNPhood TCGAbiolinks
WARNING : 15 : biobroom bsseq Chicago genomation GenomicInteractions iGC ImmuneSpaceR metaX MSnID MSstats paxtoolsr Pviz r3Cseq RiboProfiling scater
NOTE : 27
OK : 3
TOTAL : 65 / 65
RUNNING : 0
So, our next task is to make Jan’s result on docker match mine. I can’t quite remember how I got all these packages to pass locally for me. In some cases I needed to find and install Ubuntu libraries and I tried my best to keep a note of them at the time here here . Another case is that lava suggests mets but mets depends on lava. We currently solve chicken-or-egg situations manually, one-by-one. A third example is that permissions of /tmp seem to be different on docker which at least one package appears to test and depend on. We have tried changing TEMPDIR from /tmp to ~/tmp to solve that and will wait for the rerun to see if that worked. I won’t be surprised if it takes a week of elapsed time to get our results to match. That’s two man weeks of on-and-off time as we fix, automate-the-fix and wait to see if the rerun works. And this is work after data.table has already made it to CRAN; to make next time easier and less of a barrier-to-entry to start.
The point is, all this takes time behind the scenes. I’m sure other package maintainers have similar issues and have come up with various solutions. I’m aware of devtools::revdep_check, used it gratefully for some years and thanked Hadley for it in this tweet . But recently I’ve found it more reliable and simpler to run R CMD check at the command line directly using the unix parallel command. Thank you to R-core and CRAN maintainers for keeping CRAN going in 2016. There must be much that nobody knows about. Thank you to the package maintainers that use data.table and have received my emails and fixed their warnings or errors (users will never know that happened). Sorry I myself didn’t keep data.table cleaner, faster. We’re working to improve that going forward.