There is a new minor release of H2O that introduces two useful improvements to our XGBoost integration: interaction constraints and feature interactions.
Interaction Constraints
Feature interaction constraints allow users to decide which variables are allowed to interact and which are not.
Potential benefits:
- Better predictive performance from focusing on interactions that work – whether through domain-specific knowledge or algorithms that rank interactions
- Less noise in predictions; better generalization
- More control given to the user on what the model can fit. For example, the user may want to exclude some interactions even if they perform well due to regulatory constraints
(Source: https://xgboost.readthedocs.io/en/latest/tutorials/feature_interaction_constraint.html )
The H2O documentation is available here .
XGBFI-like Tool for Revealing Feature Interactions
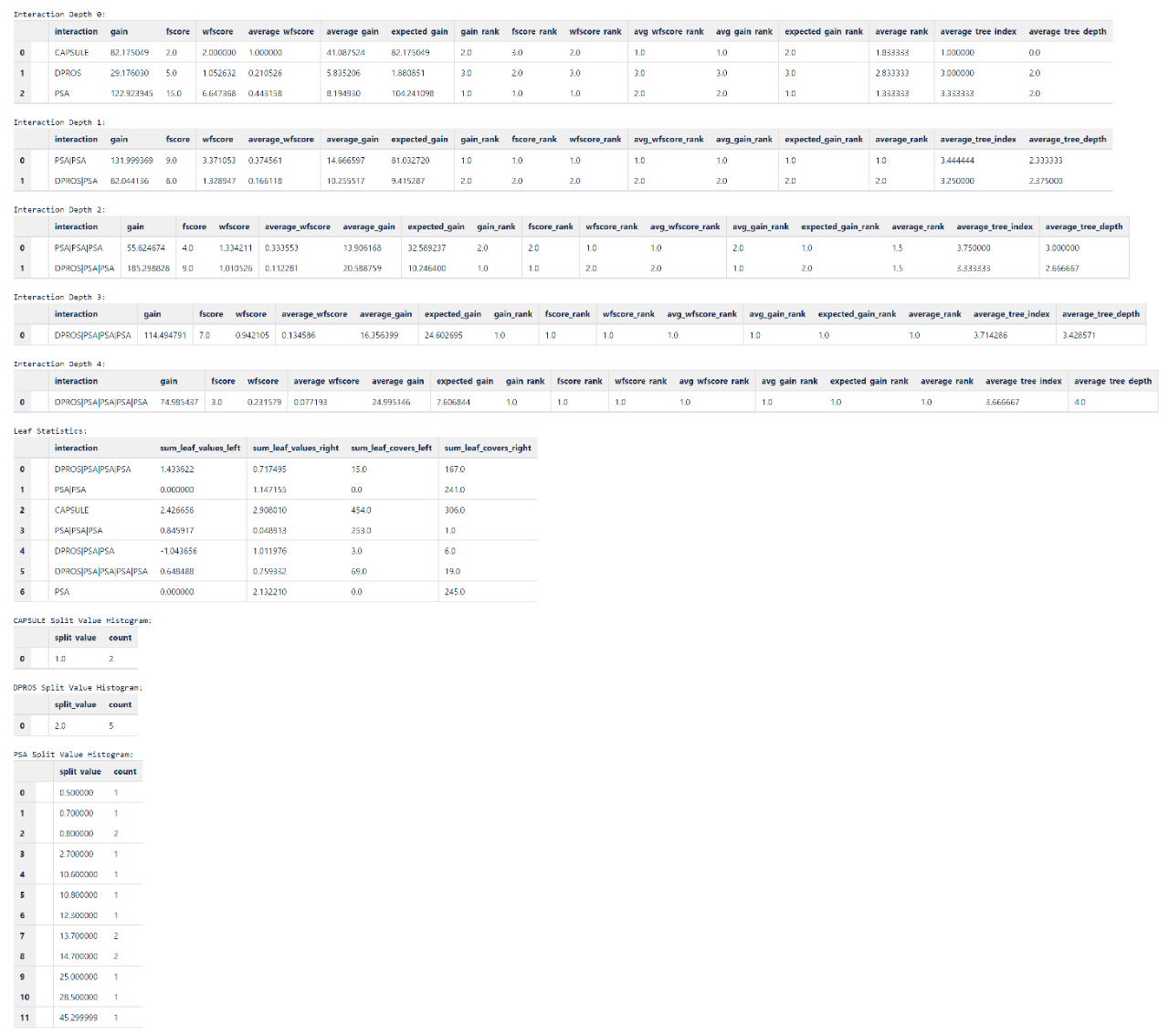
We have implemented ranks of features and feature interactions by various measures in XGBFI style. Thanks to this tool, H2O provides insights into higher-order interactions between features in trees all in a user-friendly manner. Additionally, leaf statistics and split value histograms are provided. The measures used are either one of:
Gain implies the relative contribution of the corresponding feature to the model calculated by taking each feature’s contribution for each tree in the model. A higher value of this metric when compared to another feature implies it is more important for generating a prediction.
Cover is a metric to measure the number of observations affected by the split. Counted over the specific feature it measures the relative quantity of observations concerned by a feature.
Frequency (FScore) is the number of times a feature is used in all generated trees. Please note that it does not take the tree-depth nor tree-index of splits a feature occurs into consideration, neither the amount of possible splits of a feature. Hence, it is often suboptimal measure for importance or their averaged / weighed / ranked alternatives.
The H2O documentation is available here .
Example
The Jupyter notebook demo with all example codes presented below is available here .
Train XGBoostEstimator with interaction_constraints parameter:
# start h2o
import h2o
h2o.init()
from h2o.estimators.xgboost import *
# check if the H2O XGBoostEstimator is available
assert H2OXGBoostEstimator.available() is True
# import data
data = h2o.import_file(path = "../../smalldata/logreg/prostate.csv")
x = list(range(1, data.ncol-2))
y = data.names[len(data.names) - 1]
ntree = 5
h2o_params = {
'eta': 0.3,
'max_depth': 3,
'ntrees': ntree,
'tree_method': 'hist'
}
# define interactions as a list of list of names of colums
# the lists defines allowed interaction
# the interactions of each column with itself are always allowed
# so you cannot specified list with one column e.g. ["PSA"]
h2o_params["interaction_constraints"] = [["CAPSULE", "AGE"], ["PSA", "DPROS"]]
# train h2o XGBoost model
h2o_model = H2OXGBoostEstimator(**h2o_params)
h2o_model.train(x=x, y=y, training_frame=data)The result:
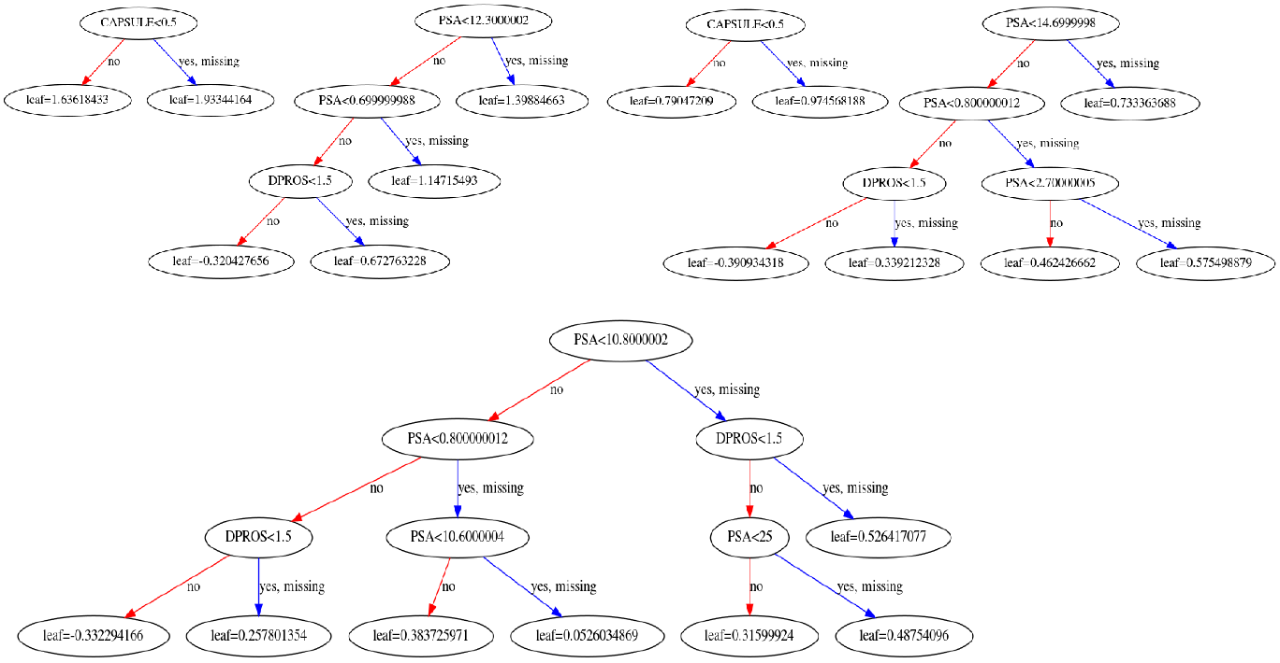

Display feature interactions:
# calculate multi-level feature interactions
h2o_model.feature_interaction()

Credits
This new H2O release is brought to you by Veronika Maurerova , Zuzana Olajcova , and Hannah Tillman .
How to Get Started?
Download H2O-3 from here and follow the steps in this example notebook . You can also check out our training center for both self-paced tutorials and instructor-led courses.