Parkinson’s disease, a condition affecting movement, cognition, and sleep, is escalating rapidly. By 2037, it is projected that around 1.6 million U.S. residents will be confronting this disease, resulting in significant societal and economic challenges. Studies have hinted that disruptions in proteins or peptides could be instrumental in the disease’s onset and progression. Consequently, delving deeper into these biological elements may provide valuable insights, potentially paving the way toward halting disease progression or even finding a cure.

Participants were assigned the mission of utilizing protein and peptide data from Parkinson’s patients to project disease progression, which could potentially spotlight pivotal molecules undergoing change as the disease evolves. This competition forms part of a larger AMP®PD initiative, a cooperative endeavor across various sectors, to identify and validate critical biomarkers for Parkinson’s, bolstering the global fight against this debilitating neurological disorder.
The Competition
As stated above, the competition’s objective was to predict the progression of Parkinson’s disease(PD) using data on protein abundance. More specifically, the participants had to predict MDS-UPDR scores, which indicate the progression of Parkinson’s disease in patients. MDS-UPDRS is a scale used to evaluate various symptoms, including both movement-related and non-movement-related symptoms, that are associated with Parkinson’s disease.
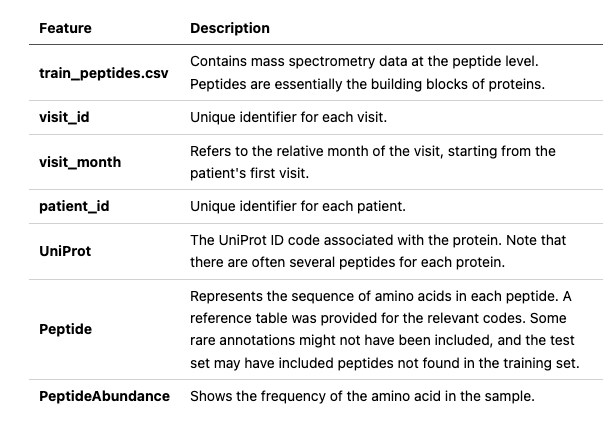
The dataset was primarily composed of protein abundance levels obtained from mass spectrometry readings of cerebrospinal fluid (CSF) samples collected from hundreds of patients over several years. This competition was unique in that it was a time-series code contest, where predictions were made using Kaggle’s time-series API on provided test set data. The data used in the competition was organized as follows:
A Notable Win: Overcoming Complexities
Against this challenging backdrop, Dmitry Gordeev, a Kaggle Competition Grandmaster and Director of Data Science & Product at H2O.ai collaborated with Konstantin Yakovlev, Head of Data Science at Palta, to clinch a remarkable win in this competition.
Dmitry’s and Konstantin’s final solution involved combining two models — a LightGBM model(LGBM) and a Neural Network (NN), by taking a simple average. These models were trained on the same set of features, including visit month, forecast horizon, target prediction month, and indicators related to patient visits and supplementary data. Interestingly, the winning solution disregarded the results of blood tests as none of their approaches or models could find significant signals in that data. Instead, the final models were trained using only the clinical and supplementary datasets.
For the LGBM, they took an alternative approach by treating it as a classification model with 87 target classes. Post-processing techniques were applied to minimize the SMAPE+1 metric.

On the other hand, the NN model utilized a simple multi-layer feed-forward architecture with a regression target and used SMAPE+1 as the loss function. They incorporated a leaky RELU activation in the last layer to handle negative predictions.

Various cross-validation schemes were explored, and they settled on leave-one-(patient)-out cross-validation — a group k-fold cross-validation with a fold for each patient. This helped to eliminate dependence on random numbers.

The full code of the winning notebook can be found here, while an elaborate solution provided by the team can be found here.
AI for Good: Impact Beyond the Competition
While the primary objective of the competition was to discover molecular indicators of Parkinson’s disease progression, the winning solution shed light on the limitations of traditional blood tests in providing the necessary information. Nonetheless, this valuable insight suggests the need for either refining the current approach or exploring alternative setups that can uncover subtle yet significant markers of disease progression.
We are immensely proud of our colleague for his achievement and commitment to making a difference reminding us that our mission of using AI for Good is not only noble but absolutely achievable.
Stay tuned for more inspiring stories from our AI for Good initiative as we continue this exciting journey of discovery and impact.